This page is in progress
This document describes the process of creation of used in the
project IDEEA external (extra) datasets, stored in Zenodo
repository (link).
library(IDEEA)
# devtools::load_all(".")
library(tidyverse)
library(data.table)
# pak::pkg_install("energyRt/merra2ools")
library(merra2ools)
library(ggthemes)
library(glue)Weather data and capacity factors
MERRA-2
For the sample from MERRA-2 dataset, it is not
ideea_sf <- get_ideea_map(nreg = 32, offshore = T, islands = T)
ideea_locid <- get_locid(ideea_sf)
n_locid <- length(ideea_locid)
ideea_locid_sf <- get_merra2_grid(type = "poly", locid = ideea_locid) |>
select(locid) |>
left_join(merra2ools::mean_cf) |>
st_make_valid()
# rename variables for consistency
names(ideea_locid_sf) <- str_replace(names(ideea_locid_sf), "waf", "wcf_") |>
str_replace("saf", "scf")
names(ideea_locid_sf)
if (F) {
# the following files will be saved in 'ideea_extra' folder
# ideea_locid
save(ideea_locid, file = ideea_extra(subdir = "merra2",
filename = "ideea_locid.RData"))
# ideea_locid_sf
save(ideea_locid_sf,
file = ideea_extra(subdir = "merra2", filename = "ideea_locid_sf.RData"))
}
ggplot() +
geom_sf(data = ideea_sf, fill = alpha("grey", .5), color = "darkgrey") +
geom_sf(data = ideea_locid_sf, fill = NA, color = "red") +
labs(title = glue("MERRA-2 grid, {n_locid} cells (locations) in total")) +
theme_bw()
d <- pivot_longer(ideea_locid_sf, cols = wcf_50m:scf_td,
names_to = "tech_type", values_to = "mean_cf") |>
select(locid, tech_type, mean_cf, geometry) |> st_as_sf()
ggplot() +
geom_sf(aes(fill = mean_cf), data = d, color = NA) +
geom_sf(data = ideea_sf, fill = NA, color = alpha("black", .25)) +
facet_wrap(~tech_type) +
scale_fill_viridis_c(option = "H") +
# labs(title = glue("MERRA-2 grid, {n_locid} cells (locations) in total")) +
theme_bw()The next step is to acquire weather data from MERRA-2 dataset,
time-series for every location (cell) that will be further used to
estimate capacity factors for wind and solar generators by hours. We use
merra2ools package to streamline the process. The weather
data available from 1980 to 2020. In this example we sample locations
for 2019, that is embedded in the IDEEA R-package. Other years are
available for download from IDEEA’s Zenodo repository, and can be
attached using ideea_extra() function.
overwrite <- FALSE
for (wyear in seq(2020, 1981, -1)) {
fname <- ideea_extra("merra2", glue("merra_raw_{wyear}.fst")) # file to save
# fpath <- ideea_extra("merra2", fname) # file name with path
if (file.exists(fname) & !overwrite) {
message("File already exists: \n ", fname)
next
}
merra <- get_merra2_subset(
locid = ideea_locid,
from = fDate(wyear, 01, 01, 0),
to = fDate(wyear, 12, 31, 23),
tz = "Asia/Kolkata"
)
# merra <- merra |> select(UTC, locid, W10M, W50M, SWGDN, ALBEDO)
message("Saving 'raw' data: ", fname)
fst::write_fst(merra, path = fname, compress = 100)
cat("File size:", {file.size(fname) |> gdata::humanReadable()}, "\n"
}Wind sites
Global Wind Atlas (GWA)
library(terra)
library(globalwindatlas)
gwa_set_dir(ideea_extra("gwa"))
gwa_get_dir()
gwa_tif <- gwa_get_wind_cf("IND", IEC = 2) # wind-class #2
## plot
gwa_rast <- rast(gwa_tif)
plot(gwa_rast, main = "Wind capacity factors (GWA)")Grouping locations by capacity factor
for (nreg in c(5, 32)) {
ob_name <- glue("gwa_iec2_r{nreg}_sf")
fname <- ideea_extra("gwa", "{ob_name}.RData") |> glue()
if (file.exists(fname)) next
ob <- gwa_group_locations(
gwa_tif = gwa_tif,
gis_sf = get_ideea_map(nreg = nreg, offshore = T, islands = T,
aggregate = T, rename = F),
aggregate_tif = 0,
drop_crumps = 100,
simplify = 0.001,
buffer = 0,
int = seq(0, 1, by = .1),
verbose = T
)
assign(ob_name, ob); rm(ob)
message("Saving ", fname)
save(list = ob_name, file = fname)
rm(fname)
}
ggplot() +
geom_sf(color = "grey30", fill = "grey", data = ideea_sf) +
geom_sf(aes(fill = eq), color = NA, data = gwa_iec2_r5_sf) +
scale_fill_viridis_d(option = "H", direction = 1, name = "CF") +
theme_bw() +
labs(title = "Wind capacity factors by level, source: Global Wind Atlas (GWA)")
# ggsave("gwa_iec2_sf.png", path = ideea_extra("gwa", check = F),
# height = 5, width = 5, scale = 1.5)Select locations with high wind potential
# parameters:
win_gwa_cf_min <- 0.2 # set lowest capacity factor (GWA)
win_merra_cf_min <- 0.2 # set lowest capacity factor (MERRA2/merra2ools)
win_onshore_MW_km2 <- 4 # maximum MW per km2 for onshore wind (assumption)
win_offshore_MW_km2 <- 4 # maximum MW per km2 for offshore wind (assumption)
for (nreg in c(5, 32)) {
ob_name <- glue("locid_win_r{nreg}_sf")
fname <- ideea_extra("merra2", "{ob_name}.RData") |> glue()
gwa_ob_name <- glue("gwa_iec2_r{nreg}_sf")
ob <- ideea_locid_sf |> # MERRA grid with average CFs (no regions)
select(-starts_with("scf_")) |> # drop solar cf-data
st_make_valid() |>
st_intersection( # find an intersection between GWA and MERRA sf-objects
filter(
# drop low-potential sites, based on both GWA and MERRA
get(gwa_ob_name), # GWA sf object for n-region case
int >= win_gwa_cf_min # GWA group (see above)
),
dimensions = c("polygon")
) |>
filter(
wcf_100m >= win_merra_cf_min # MERRA
) |>
st_make_valid() |>
mutate(
# estimate surface area of each geometry and maximum potential in MW
area = units::set_units(st_area(geometry), "km^2"),
MW_max = as.numeric(
round(
if_else(
offshore, # or us grepl("off", reg_off),
win_offshore_MW_km2 * area, # area-based estimate, max onshore MW
win_onshore_MW_km2 * area # area-based estimate, max offshore MW
),
0)
),
.before = "geometry"
)
assign(ob_name, ob); rm(ob)
message("Saving ", fname)
save(list = ob_name, file = fname)
rm(fname)
}Cluster locations
# make a sample of capacity factors time-series for clustering
merra <- fst::read_fst(ideea_extra("merra2", "merra_raw_2019.fst"),
as.data.table = TRUE)
# estimate capacity factors at different hub-heights
merra_wind <- merra |>
# fPOA() |> # solar (POA)
fWindCF(50, return_name = "wcf_50m") |> # wind
fWindCF(100, return_name = "wcf_100m") |>
fWindCF(150, return_name = "wcf_150m") |>
select("UTC", "locid", starts_with("wcf_"))
# cluster locations based on
for (nreg in c(5, 32)) {
# for (nreg in c(32)) {
regN <- glue("reg{nreg}")
regN_off <- glue("reg{nreg}_off")
ob_name <- glue("locid_win_cl_r{nreg}")
ob_sf_name <- glue("{ob_name}_sf")
fname <- ideea_extra("merra2", glue("locid_win_cl_r{nreg}.RData"))
fname_sf <- str_replace(fname, "\\.RData", "_sf\\.RData")
# load sf-object (map) of filtered for wind locations with MERRA2 grid
locid_win_sf <- ideea_extra("merra2", glue("locid_win_r{nreg}_sf.RData")) |>
load()
# cluster locations for each group (region),
# as the results, cluster # will be assigned for each locid by region and k
# where k is the number of clusters in region, from 1:N
# N - number of MERRA2-cells in region
ob <- cluster_locid(
merra_wind,
varname = "wcf_100m",
# locid_info = get(locid_win_sf),
locid_info = filter(get(locid_win_sf), int == win_merra_cf_min), # cf layer
group = regN_off,
weight = "MW_max",
max_loss = 0.,
# k is the number of clusters to consider. k <= N
# For large regions with many locations, the clustering process can be long.
# We can limit the options with a give sequence:
k = c(1:20, 25, 30, 40, 50, 75, 100, 150, 200, 300, 500, 1000),
plot = T,
verbose = T
)
# add 'reg{nreg}' column
if (is.null(ob[[regN]])) {
ob <- ob |>
mutate(
"{regN}" := str_replace_all(get(regN_off), "_off", ""), .before = 1
)
}
# rename 'ob' and save
assign(ob_name, ob); rm(ob)
message("Saving clustering results:")
cat(" ", fname, "\n")
save(list = ob_name, file = fname)
# convert cluster-table to sf-object (map), adding geometry for each cell
ob_sf <- get(locid_win_sf) |>
# select(-starts_with("scf_")) |>
st_make_valid() |>
select(-MW_max) |>
right_join(get(ob_name), relationship = "many-to-many") |>
filter(!is.na(cluster)) |>
mutate(cluster = factor(cluster)) |>
st_as_sf()
assign(ob_sf_name, ob_sf); rm(ob_sf)
cat(" ", fname_sf, "\n")
save(list = ob_sf_name, file = fname_sf)
}Solar sites
# parameters:
# sol_cf_min <- 0.1 # set lowest capacity factor (MERRA2/merra2ools)
sol_onshore_MW_km2 <- 80 # assumption (see info), maximum MW per km2
sol_offshore_MW_km2 <- 10 # assumption (see info), maximum MW per km2
for (nreg in c(5, 32)) {
ob_name <- glue("locid_sol_r{nreg}_sf")
fname <- ideea_extra("merra2", "{ob_name}.RData") |> glue()
ob <- ideea_locid_sf |> # MERRA grid with average CFs (no regions)
select(-starts_with("wcf_")) |> # drop solar cf-data
st_make_valid() |>
st_intersection(
get_ideea_map(nreg, offshore = T, islands = T, rename = F),
dimensions = c("polygon")
) |>
st_make_valid() |>
mutate(
# estimate surface area of each geometry and maximum potential in MW
area = units::set_units(st_area(geometry), "km^2"),
MW_max = as.numeric(
round(
if_else(
offshore, # or us grepl("off", reg_off),
sol_offshore_MW_km2 * area, # area-based estimate, max onshore MW
sol_onshore_MW_km2 * area # area-based estimate, max offshore MW
),
0)
),
.before = "geometry"
)
assign(ob_name, ob); rm(ob)
message("Saving ", fname)
save(list = ob_name, file = fname)
rm(fname)
}Cluster locations
# make a sample of capacity factors time-series for clustering
merra <- fst::read_fst(ideea_extra("merra2", "merra_raw_2019.fst")) |>
as.data.table()
# estimate capacity factors at different PV-tracking systems by location
merra_solar <- merra |>
fPOA(array.type = c("fh", "fl",
# "th", "tv", # rarely used & similar to other types
"tl", "td")) |> # solar (Plain of Array irradiance)
mutate(
# simplified version of capacity factors,
# assuming pick of output when POA >= 1000 Watt/m^2
scf_fh = round(POA.fh / 1e3, 3),
scf_fl = round(POA.fl / 1e3, 3),
# scf_th = round(POA.th / 1e3, 3),
# scf_tv = round(POA.tv / 1e3, 3),
scf_tl = round(POA.tl / 1e3, 3),
scf_td = round(POA.td / 1e3, 3)
) |>
mutate(
# curtail cf > 1 (when POA > 1kW/m^2)
scf_fh = if_else(scf_fh > 1, 1, scf_fh),
scf_fl = if_else(scf_fl > 1, 1, scf_fl),
# scf_th = if_else(scf_th > 1, 1, scf_th),
# scf_tv = if_else(scf_tv > 1, 1, scf_tv),
scf_tl = if_else(scf_tl > 1, 1, scf_tl),
scf_td = if_else(scf_td > 1, 1, scf_td)
) |>
select(locid, UTC, starts_with("scf_"))
summary(merra_solar)
merra_solar
# locid_sol_sf <- ideea_locid_sf |>
# st_make_valid() |>
# st_intersection(st_make_valid(ideea_sf)) |>
# st_make_valid() |>
# mutate(
# area = units::set_units(st_area(geometry), "km^2"),
# offshore = if_else(grepl("off", reg_off), T, F),
# MW_max = as.numeric(round(if_else(
# offshore,
# sol_offshore_MW_km2 * as.numeric(area),
# sol_onshore_MW_km2 * as.numeric(area)), 0)),
# .before = "geometry"
# )
# cluster locations based on temporal correlation
for (nreg in c(5, 32)) {
regN <- glue("reg{nreg}")
regN_off <- glue("reg{nreg}_off")
ob_name <- glue("locid_sol_cl_r{nreg}")
ob_sf_name <- glue("{ob_name}_sf")
fname <- ideea_extra("merra2", glue("locid_sol_cl_r{nreg}.RData"))
fname_sf <- str_replace(fname, "\\.RData", "_sf\\.RData")
# load sf-object (map) of filtered for solar locations with MERRA2 grid
locid_sol_sf <- ideea_extra("merra2", glue("locid_sol_r{nreg}_sf.RData")) |>
load()
# cluster locations by group (region),
# as the results, cluster # will be assigned for each locid by region and k
# where k is the number of clusters in region, from 1:N
# N - number of MERRA2-cells in region
ob <- cluster_locid(
merra_solar,
varname = "scf_tl",
locid_info = get(locid_sol_sf),
group = regN_off,
weight = "MW_max",
max_loss = 0.,
# k is the number of clusters to consider. k <= N
# For large regions with many locations, the clustering process can be long.
# We can limit the options with a give sequence:
k = c(1:20, 25, 30, 40, 50, 75, 100, 150, 200, 300, 500, 1000),
plot = T,
verbose = T
)
# add 'reg{nreg}' column
if (is.null(ob[[regN]])) {
ob <- ob |>
mutate(
"{regN}" := str_replace_all(get(regN_off), "_off", ""), .before = 1
)
}
# rename 'ob' and save
assign(ob_name, ob); rm(ob)
message("Saving clustering results:")
cat(" ", fname, "\n")
save(list = ob_name, file = fname)
# convert cluster-table to sf-object (map), adding geometry for each cell
ob_sf <- get(locid_sol_sf) |>
# select(-starts_with("scf_")) |>
st_make_valid() |>
select(-MW_max) |>
right_join(get(ob_name), relationship = "many-to-many") |>
filter(!is.na(cluster)) |>
mutate(cluster = factor(cluster)) |>
st_as_sf()
assign(ob_sf_name, ob_sf); rm(ob_sf)
cat(" ", fname_sf, "\n")
save(list = ob_sf_name, file = fname_sf)
}Getting capacity factors
# default tol
get_ideea_cf(resource = "sol", nreg = 5, year = 2019)
get_ideea_cf(resource = "sol", nreg = 32, year = 2019)
get_ideea_cf(resource = "win", nreg = 5, year = 2019)
get_ideea_cf(resource = "win", nreg = 32, year = 2019)
# tol = 1%
get_ideea_cf(resource = "sol", nreg = 5, tol = 0.01, year = 2019)
get_ideea_cf(resource = "sol", nreg = 32, tol = 0.01, year = 2019)
get_ideea_cf(resource = "win", nreg = 5, tol = 0.01, year = 2019)
get_ideea_cf(resource = "win", nreg = 32, tol = 0.01, year = 2019)Visualizing
Functions ideea_snapshot_cf and
ideea_gif_sf designed to visualize capacity factors for
wind and solar generators. The first function allows to plot an instance
of potential generation for a given hour (timslice) for a particular
cluster. The second function creates a gif-file with a sequence of
timeslices.
Solar
resource <- "sol"; cf_name <- "scf_tl"
nreg <- 5
tol <- 0.01
# shape files
ideea_sf <- get_ideea_map(nreg = nreg, offshore = T, islands = T)
ideea_cl_sf <- get_ideea_cl_sf(resource = resource, tol = tol)
# total clusters (maximum across regions)
ideea_cl_sf$cluster |> unique()
# clusters' plot
plot(ideea_cl_sf["cluster"])
# capacity factors
x <- get_ideea_cf(resource, tol = tol, nreg = nreg, year = 2019)
# plot a snapshot for a (random) slice
ideea_snapshot_cf(x, ideea_cl_sf, ideea_sf, cf_name = cf_name)
# create a gif-file
# pull slices from partial calendar
calendar_1day_per_month <-
ideea_modules$electricity$reg7_base$partial_calendar_1day_per_month
slices_1day_per_month <- calendar_1day_per_month@timetable$slice
# make a gif
ideea_gif_cf(x, ideea_cl_sf, ideea_sf, cf_name = cf_name,
slice = slices_1day_per_month,
fps = 5, gif.width = 864, gif.height = 864,
filename = glue("{resource}.gif"))
Solar capacity factors, animated
Wind
Similarly, we can visualize capacity factors for wind generators.
resource <- "win"; cf_name <- "wcf_100m"
nreg <- 5
tol <- 0.05
# ideea_sf <- get_ideea_map(nreg = nreg, offshore = T, islands = T)
ideea_cl_sf <- get_ideea_cl_sf(resource = resource, tol = tol)
x <- get_ideea_cf(resource, tol = tol)
# plot
ideea_snapshot_cf(x, ideea_cl_sf, ideea_sf, cf_name = cf_name)
# gif
ideea_gif_cf(x, ideea_cl_sf, ideea_sf, cf_name = cf_name,
slice = slices_1day_per_month,
fps = 5, gif.width = 864, gif.height = 864,
filename = glue("{resource}.gif"))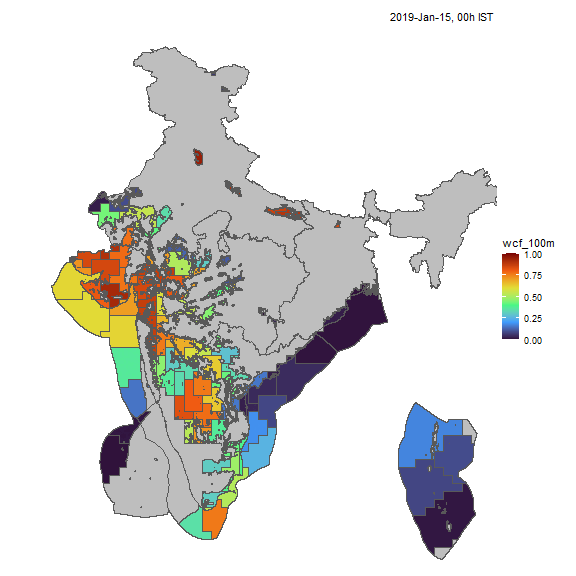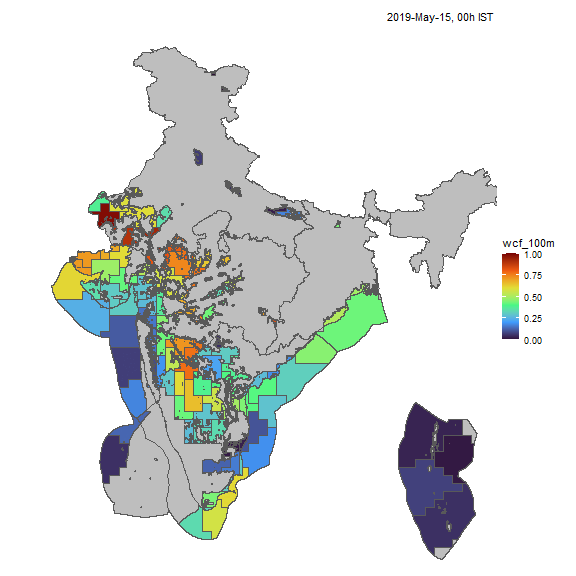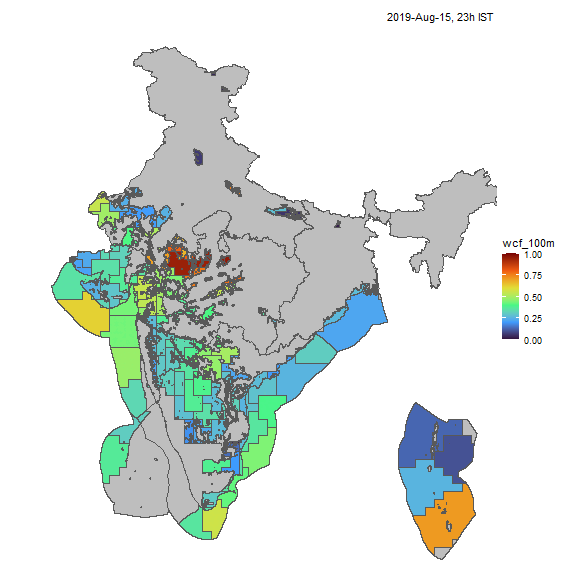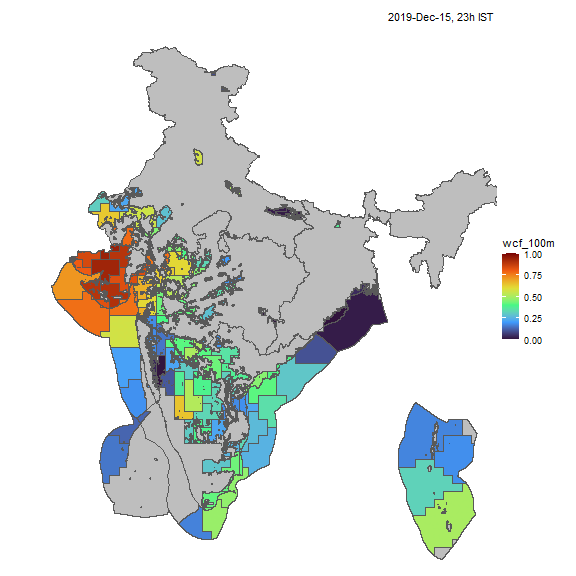
Wind capacity factors, animated
tbc…
